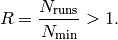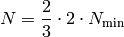Creating run combination files – prof-runcombs¶The prof-runcombs script assists you with the creation of MC-run-combinations, i.e. the MC runs that are used as anchor points for the polynomial parameterization of the MC response function and with the extraction of (a subset of) the run combinations from a result list file. The mode of operation is given as first argument create or extract. If no mode is given create mode is assumed. In create mode the location of the MC runs can be specified by --mcdir MCDIR or --datadir DATADIR, as for the main prof-interpolate and prof-tune scripts. The combinatorics of the run combinations can be chosen with (multiple) -c COMBINATION. In extract mode the file to extract run combinations from must be given as second argument. Runs can be excluded in both modes by two different ways: In create mode only the runs that fulfil the above criteria, i.e. are not excluded and are within the parameter range (if both options are used), are used to sample the run combinations from. In extract mode only those run combinations that do not contain any run that is excluded are extracted. If -r RANGEFILE is used to exclude runs in extract mode, the location of the MC runs must be given by --mcdir MCDIR. See Run combination file format for details of the output run combination file syntax. Oversampling ratio¶Todo Put this section into a more general user guide The coefficients of the polynomial interpolations are calculated by solving a system of linear equations. The minimial number of runs is given by the number of parameters that are tuned and the order of the polynomial that is used for the interpolations:
As discussed in this paper (arxiv:0907.2973) it is in principle possible to use the minimal number of anchor points. But this would tie the interpolation strictly to the anchor points and we know that the MC response function in each bin is more complex than the polynomials we use. To take the incapabilities of the polynomials into account we advise to use a higher number of anchor points. That is the ratio of the number of runs over the minimal number is larger than 1,
Professor then uses a singular value decomposition to calculate the coefficients resulting in a least squares fit. In fact we found that an oversampling ration of 2 is advisable while values above 3.5 do not necessarily lead to better results. Second, it is important to check that the results one gets with Professor do not depend on the specific choice of anchor points i.e. the results should be robust against using different sets of anchor points. The way to do this with Professor is to create different subsets of anchor points that are used for the interpolations. Usually we use 100 such subsets. Idealy, these subsets are disjunct. This is not feasable due to the high number of minimal anchor points one usually needs. Instead a certain level of overlapping between these subsets is accepted and in conclusion one has to find a compromise between the level of overlap between the subsets and a reasonable oversampling ratio. A good rule of thumb is to generate
runs. Todo Calculate overlap probability. Examples¶prof-runcombs create --mcdir mc/ -c 0:1 -c 30:100 -o mycombinations
This will create a file mycombinations. 101 run combinations are generated. One run combination will contain all runs, and 100 combinations will contain #(all runs) - 30. prof-runcombs extract mycombinations --mcdir mc/ -r narrow.ranges -o mynarrowcombinations
This will create mynarrowcombinations that will consist of all the run combinations from mycombinations that use only MC runs within the ranges given in narrow.ranges. Command-line options¶The --datadir DATADIR and related options are used as normal to specify the reference data, MC runs, and interpolation objects: see the path options page.
|
||||||||||||||||||||||||||||||||||||